

Protein-protein interactions by single-molecule methods
 Single-molecule fluorescence methods have provided many new insights into the biological processes dominated by macromolecules. In contrast to ensemble-averaged measurements, single-molecule measurements not only describe the real-time conformational dynamics of individual molecules but also identify details of protein–protein or protein–environment interactions that are undetected in ensemble-averaged experiments.
Single-molecule fluorescence methods have provided many new insights into the biological processes dominated by macromolecules. In contrast to ensemble-averaged measurements, single-molecule measurements not only describe the real-time conformational dynamics of individual molecules but also identify details of protein–protein or protein–environment interactions that are undetected in ensemble-averaged experiments.
In vitro single-molecule methods are feasible for identifying the key points in signaling pathways because in vitro experiments suppress the long-distance transportation of signal transduction components between different organelles in living cells and restrict the target motion range near the slide surface. The most common in vitro single-molecule assays are usually performed using conventional total internal reflection fluorescence microscopy (TIRFM).
Reference:“Reactive oxygen species‐mediated BIN 2 activity revealed by single‐molecule analysis.” Song, S., Wang, H., Sun, M., Tang, J., Zheng, B., Wang, X., and Tan, Y.New Phytol. 223, 692–704 (2019).“Single-Molecule Fluorescence Methods to Study Plant Hormone Signal Transduction Pathways,” S. Song, J. Chang, C. Ma, and Y.-W. Tan,Front. Plant Sci., 02 (2017).
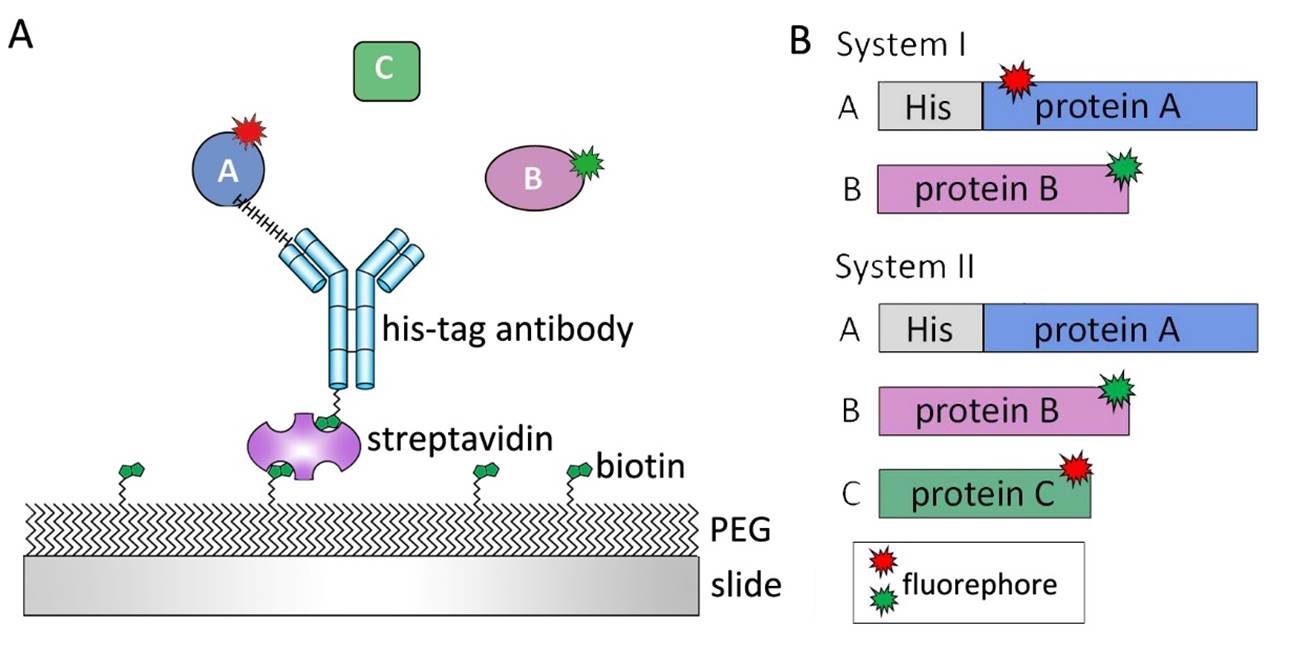
Figure 1Single-molecule experimental design. (A) Cleaned slides are passivated by poly-ethylene glycol (PEG) and biotin-PEG to eliminate unspecific protein bindings. Protein A was immobilized on slide surface by biotin-streptavidin–antibody linker. Add Protein B or even protein C on slide and observe their interaction on real-time. (B) For two protein system, one protein with his tag immobilized on slide surface. Both of them should be labelled by fluorophores. For three protein system, protein A which can connect protein B and protein C was designed with His-tag to be immobilized on slide surface. To observe interactions between three proteins, at least protein B and C should be labelled.